While methane (CH4) is a potent greenhouse gas that contributes to climate change, the biological transformation of CO2 into methane also holds great promise as a renewable energy source. Understanding the fundamental mechanisms behind methane formation could lead to advancements in sustainable energy technologies and environmental conservation.
At the heart of biological methane production – the methanogenesis – sits the enzyme MCR with its unique nickel complex F430. In order to catalyze methane production, F430 must be reduced, which is one of the most challenging redox reactions in nature. It has long remained an open question how early life forms could conduct strongly reducing electrons into the enzyme.
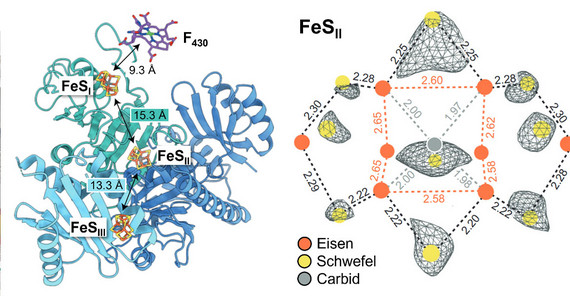
In their study, the research team succeeded in isolating and characterizing the MCR activation complex from the model archaeon Methanococcus maripaludis. Methanogenic archaea are microorganisms that have existed for billions of years, producing up to one billion tons of methane annually. The new electron microscopy structure now suggests that the MCR activation complex contains three uniquely coordinated and highly specialized redox cofactors that were previously thought to be exclusive to nitrogenase – an enzyme complex that is responsible for nitrogen fixation in living organisms. “The spectroscopy yielded the final piece of evidence that the cofactors are comprised of iron and sulfur”, explains Sven T. Stripp, co-author from University of Potsdam. Jan Schuller, the study’s senior author, adds: “This striking similarity suggests that, despite performing entirely different functions, these systems share an evolutionary relationship.” He concludes: “Ultimately, our study establishes an unprecedented evolutionary connection between two fundamental biological processes: methanogenesis and nitrogen fixation.”
Methanogenesis is a process that dates back to the earliest history of life on Earth, evolutionary even predating photosynthesis. It is not only responsible for methane emissions but also forms the foundation for other metabolic networks crucial for life. A deeper understanding of these mechanisms advances both our fundamental knowledge of molecular evolution and its possible biotechnological applications, ultimately aiming to mitigate methane emissions.
Image 1: Team photo (left 2nd: Sven T. Stripp). Credit: Tomas Pascoa
Image 2: Electron microscopy structure of the MCR activation complex with three novel iron-sulfur cofactors (FeS). Credit: Sven T. Stripp
Link to Publication: Fidel Ramírez-Amador, Sophia Paul, Anuj Kumar et al. Sven T. Stripp & Jan M. Schuller, Structure of the ATP driven Methyl-coenzyme M reductase activation complex. Nature (2025). https://www.nature.com/articles/s41586-025-08890-7
Link to Press Release University of Marburg:https://www.uni-marburg.de/de/aktuelles/news/2025/methan25
Contact:
Dr. Sven T. Stripp
Institute of Chemistry, University of Potsdam
E-Mail: sven.strippuuni-potsdampde
Phone: +49 331 977-5236