SynBio Tools for Strain Engineering
The transition to a bio-based economy demands organisms, like plants or microorganisms, that utilize sustainable biomass to produce proteins or compounds of industrial relevance like e.g. biofuels, biopolymers, pharmaceuticals or food supplements.
This usually requires a variety of genomic manipulations, like the insertion of foreign genes necessary to utilize alternative substrates or synthesize the desired compound, or the deletion of native genes, which might negatively interfere with the foreign biochemical pathway.
Furthermore, precise tools to regulate the expression levels of the gene products are needed to optimize product yield.
In our group, we develop robust genetic tools, which help to customize host organisms in order to create improved biotechnological chassis which ideally combine optimized productivity with an easy and cost-efficient cultivation. Our initial tool development starts in the model organism Saccharomyces cerevisiae, a powerful and favored host for biotechnological purposes, due to its short generation time, the availability of methods for its cheap and easy manipulation, and its well-known genome.
As soon as a standardized workflow is established, it is transferred to other biotechnologically interesting yeasts like Pichia pastoris or Yarrowia lipolytica or even non-conventional yeast species with beneficial features.
Key publications
An Optimized Genotyping Workflow for Identifying Highly SCRaMbLEd Synthetic Yeasts
Collaborative research with the research group of Dr. Daniel Schindler.
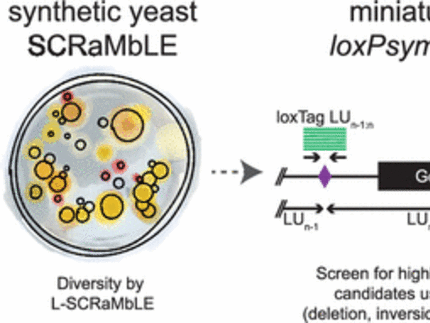
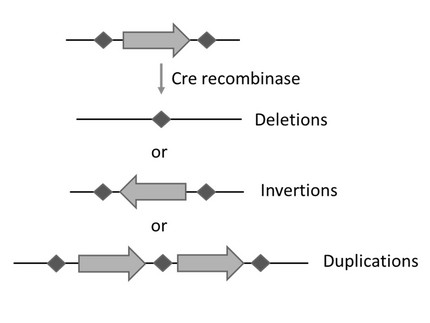
Synthetic Sc2.0 yeast strains contain hundreds to thousands of loxPsymrecombination sites that allow restructuring of the Saccharomyces cerevisiae genome by SCRaMbLE, an efficient method to generate optimized chassis for biotechnological applications. Here, we show that the light-inducible Cre recombinase L-SCRaMbLE can efficiently generate diverse recombination events when applied to synthetic Sc2.0 yeast strains. Further, we developed a set of qPCR primers for genotyping across loxPsym sites simplifies the detection of deletions, inversions and translocations on a genomic level.
PhiReX 2.0: A Programmable and Red Light-Regulated CRISPR-dCas9 System for the Activation of Endogenous Genes in Saccharomyces cerevisiae
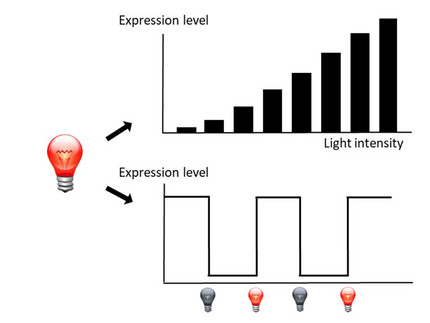
PhiReX 2.0 allows tunable and reversible red light-mediated regulation of native yeast genes without promoter manipulation at the genomic level. The CRISPR-Cas9-based design allows flexible targeting of the TF to any promoter sequence through an efficient Golden Gate-based sgRNA cloning approach.
L-SCRaMbLE as a tool for light-controlled Cre-mediated recombination in yeast
L-SCRaMbLE allows tight regulation of recombinase activity with up to 179-fold induction upon exposure to red light. The extent of recombination depends on induction time and concentration of the chromophore phycocyanobilin (PCB), which can be easily adjusted. When applied to synthetic Sc2.0 yeast strains, the red light-regulated Cre recombinase allows synthetic evolution-based optimization of host strain genomes in order to create improved chassis for specific applications.
PhiReX – a programmable and red light-regulated protein expression switch for yeast
The replacement of chemical inducers for fine-tuning or overexpression of proteins by red light-regulated transcription factors offers compelling advantages:
- Cheap and nontoxic induction system
- Easy to handle on/off switch without manipulating the cell culture
- No interference with the host metabolism
- Independency of chromophore supply
AssemblX: a user-friendly toolkit for rapid and reliable multi-gene assemblies
This assembly strategy allows webtool-supported pathway construction using homology-based cloning methods without the need for prepared part libraries.