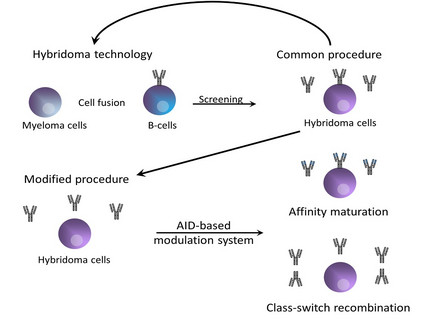
In vitro modulation of antibody affinities
Monoclonal antibodies play a crucial role in a variety of scientific assays. The functional requirements concerning their specificity as well as the affinity are quite high. A lot of antibodies for scientific purposes were generated by classical hybridoma technology. Unfortunately most of these antibodies do not fulfill the specific requirements and have to be modified in vitro by using very laborious and expensive methodology. These methods often includes the modulation of either the subclass or the affinity of the antibody.
In vivo modulation of antigen-induced immunglobulin diversification is performed by a protein called activation-induced cytodine deaminase (AID). The AID itself induces somatic hypermutations (SHM) and class-switch recombination (CSR) in activated B cells. SHM process leads to point mutations in the heavy and light chain variable gene regions especially within the complementarity determining regions (CDR) by a deamination reaction of deoxycytidine (dC) residues into deoxyuridines (dUs) yielding dU:dG mismatches. In the end a base excision repair mechanisms will be activated and causes DNA modifications that changes the affinity of an antibody. The same AID induced dC:dG mismatches occurs within the switch (S) regions of the heavy chain Ig gene loci and induces the CSR machinery by which the antibody subclass switches.
Our aim is to build up an efficient and flexible in vitro approach by using an AID based antibody affinity maturation system. This artificial system could be used to modulate antibody affinities directly in cell culture with regard to the final application.